If redirect takes more than 5 seconds, please click here.
 After years in which the Sanger sequencing has been the gold standard of molecular genetics diagnostics, Next Generation Sequencing (NGS) is going to take over. NGS is also known as high-throughput sequencing (high yield sequencing) as it allows to sequence many fragments in parallel (which was impossible by traidional Sanger sequencing).
After years in which the Sanger sequencing has been the gold standard of molecular genetics diagnostics, Next Generation Sequencing (NGS) is going to take over. NGS is also known as high-throughput sequencing (high yield sequencing) as it allows to sequence many fragments in parallel (which was impossible by traidional Sanger sequencing).There are several NGS systems, which have been developed by different companies. However, all these systems share at least three fundamental steps: DNA preparation and immobilization (i.e. the preparation of the so-called sequencing library), amplification and sequencing. Therefore, in summary, Next Generation Sequencing is made by:
- preparation of the sequencing library
- amplification
- sequencing
1. DNA PREPARATION AND IMMOBILIZITION DNA (I.E. CREATION OF SEQUENCING LIBRARY)
The DNA sample is prepared by a process of random fragmentation. The fragments obtained are added to preset sequences (known as adapters) that are necessary to anchor and immobilize the DNA to a support on which the sequencing reaction will take place. DNA fragments prepared by the addition of adapters constitute the so-called sequencing library. There are at least three different types of adapters and thus three different ways to prepare a sequencing library: linear adapters, circular adapters and bubble adapters. Naturally, there are also different types of anchor systems. In the SOLiD syste, for instance, fragments are anchored to a glass plate.
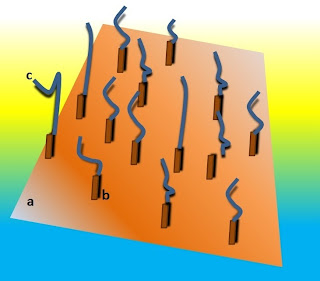 |
| Fig. 1: schematic represetantion of a sequencing library immobilized on a sequencing support: a: support; b: adapters; c: DNA fragments anchored to the support. |
2. AMPLIFICATION
Amplification can take place in emulsion (Roche and SOLiD systems) or in solution. For example, in the GS FLX system by Roche, sequencing library fragments are incorporated into a microscopic bubble of water together with the so-called enrichment beads (small balls to which adapters can bind). Amplification reaction (PCR) takes place in these aqueous microbubbles, where DNA fragments are amplified many times. Clonal copies of the fragments bind to the entire surface of the enrichment beads. Enrichment beads are then deposited on the so-called Picotiter plate, which is the support for the sequencing reaction.
3. SEQUENCING
Sequencing reaction takes place thanks to extremely complex mechanisms based on fluidic systems running on a microlitric scale, regulating the flow of reagents that react with the immobilized DNA. In the single nucleotide interrogation systems*, at each sequencing cycle immobilized DNA is mixed with a solution containing a single nucleotide (which, if complementary to the sequence, is incorporated). Then the sequencing support is washed and the DNA is mixed with another solution containing another type of nucletoide. If the nucleotide is incorpatated, the event is registered by the machine. Registration may be based on molecular imaging systems (like in the GS FLX system by Roche, for example, where the reaction of incorporation of the nucleotide into the growing complementary strand occurs in conjunction of the emission of a flash of light, hence the name of pyrosequencing, from the ancient Greek pyros, which means fire). In the Ion Torrent systems (Life Technologies) the registration of the incorportation event is based on the detection of the emission of hydrogen ions that are released during the polymerization reaction (semiconductor system).
As said at the beginning, NGS systems are also called high-throughput (high yield) because DNA fragments can be sequenced in parallel.
*In GS FLX pyrosequencing system and in the Ion Torrent system (Life Technologies), each nucleotide is interrogated individually. By contrast, in the Genome Analzyzer system (Illumina) all four nucleotides are interrogated in parallel.
To learn more about Next Generation Sequencing, Allongenetics recommend these on Amazon:
To learn more about Next Generation Sequencing, Allongenetics recommend these on Amazon:
| Next Generation Sequencing Technologies in Medical Genetics (SpringerBriefs in Genetics) |









